AI4Life Calcium Imaging Denoising Challenge 2025¶
NOTICE: Submission deadline has been extended to August 31st 2025!¶
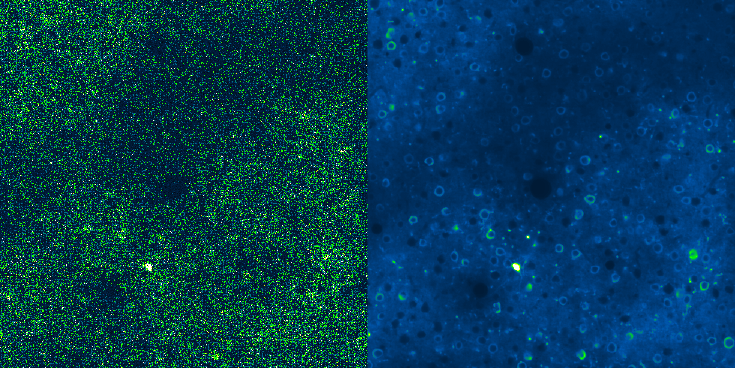
Denoising in Calcium Imaging¶
Fluorescence microscopy enables the visualization of dynamic biological processes at both cellular and subcellular scales, providing crucial insights into cellular structures, functions, and interactions of living cells.
Among the most widely used modalities is calcium imaging, which relies on genetically encoded calcium indicators (GECI) to indirectly measure intracellular calcium transients associated with cellular activity in neurons and other kinds of cells. These indicators fluoresce upon binding to calcium ions, allowing researchers to monitor functional activity across populations of living cells.
Calcium imaging data can be generally high-sensitive and bright, making it compatible with acquisition techniques such as two-photon microscopy. This allows measurements in highly scattering media, such as intact mammalian tissues, with good signal-to-noise ratios and without the need for averaging. Moreover, calcium imaging can capture the spiking activity of hundreds to thousands of neurons simultaneously, while maintaining single-cell resolution. This approach has been crucial in studying neuronal activity, synaptic transmission, and cellular signalling pathways.
Despite its advantages, calcium imaging presents several technical limitations. Its slower temporal resolution compared to other techniques such as voltage imaging makes it unsuitable for detecting fast events with high precision. As a result, while calcium imaging remains invaluable for certain appliactions, it cannot provide a full picture of neuronal electrical activity. Moreover, the acquisition of high-quality, noise-free calcium imaging data is inherently difficult in practice due to noise in the acquired images. Noise in fluorescence microscopy arises from different sources, including photon shot noise and read noise, and can significantly mask the true biological signal.
Aim of the Challenge¶
The presence of noise in calcium imaging data poses a significant issue to signal reconstruction. This motivates the development of specialized denoising methods that are specific for this setting. In particular, calcium signals are both spatially and temporally structured.
Effective denoising methods must exploit information along both dimensions to preserve the dynamics of cellular activity, with particular focus to preserve the temporal profile of the signal.
In this challenge, we highlight the importance of addressing both spatial and temporal reconstruction quality, and design evaluation metrics that explicitly quantify the denoising performances along these two dimensions.
Moreover, for a denoising method to be applicable in practice, it must generalize across heterogeneous image content (e.g, variation in morphology, fluorescence intensity, or background structure) and remain robust under different noise regimes. To address this, we designed a set of test scenarios with different noise levels and structural content.
An important limitation in denoising method research is the difficulty of obtaining clean ground truth data in real experiments. Even when acquired in higher signal-to-noise conditions, the residual noise might make it difficult to use classical metrics for reconstruction quality estimation in a rigorous way. Recognising this, we designed this challenge to focus on synthetic datasets, were the ground truth signal is exactly known and noise can be introduced in a controlled way to ensure realistic conditions. This allows for a more precise and fair quantitative evaluation of algorithm performance.
Moreover, the same limitation applies not only to evaluation but also to model training. Supervised learning approaches rely on access to clean target data, which is typically scarcely available in real fluorescence microscopy. For this reason, this challenge is open to unsupervised or self-supervised denoising methods, which do not require paired noisy-clean examples and can generalize to real-world settings.
In this challenge, we will evaluate both new and already enstablished method for imaging denoising.
Call for Evaluation Methods¶
We recognise that synthetic benchmarks cannot fully replace real experimental validation. While human expertise is crucial and effective to assess the quality of reconstruction, this approach is not feasible for automatically evaluating denoising methods. As such, this challenge also serves as a call to the community to develop and investigate evaluation methods that can operate in the absence of a clean ground truth and operate without human intervention, in particular for real calcium imaging datasets. Developing such methods is essential to better assess real-world applications of denoising methods and improve the translational impact of denoising models in neuroscience and cell biology.
About this challenge¶
This challenge is organized by the AI4Life Open Calls and Open Challenges team. We aim to showcase bioimage analysis problems to a broader audience of computational experts and work together in friendly competition, jointly pushing the boundaries of what automated solutions can do for a given analysis task.
About us¶
AI4Life is a Horizon Europe-funded project that brings together the computational and life science communities.
Its goal is to empower life science researchers to harness the full potential of Artificial Intelligence (AI) and Machine Learning (ML) methods for bioimage analysis, particularly microscopy image analysis, by providing services, and developing standards aimed at both developers and users.
AI4Life promises to create harmonized and interoperable AI tools & methods via open calls and public challenges and bring these developments to researchers via strategic outreach and advanced training.
The services provided and solutions developed within the AI4Life framework are crucial to solving today’s microscopy image analysis problems and will contribute to boosting the pace of biological and medical insights and discovery in the coming years.
The BioImage Model Zoo and FAIR data principles are core facets of the AI4Life project.

AI4Life has received funding from the European Union’s Horizon Europe research and innovation programme under grant agreement number 101057970. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.
